Phosphorylated Peptide Synthesis
Phosphorylation Post-Translational Modification (PTM)
Phosphorylation is the most important and well-studied Post-Translational Modification (PTM) in cellular activities. This reaction is involved in the regulation of many cellular processes including cell cycle, growth, apoptosis and signal transduction pathways.
Phosphorylation is controlled by kinases and phosphatases that work independently and in balance to regulate glycolysis, protein-protein interactions, protein degradation, protein inhibition, or homeostasis, among others.
This PMT is simple, flexible and reversible. Using ATP as a substrate, kinases are covalently attaching a phosphate group to the side chain of Serine (pSer), Threonine (pThr), or Tyrosine (pTyr) residues that can conversely be removed by phosphatases.
Protein phosphorylation
In the human proteome, one-third of the proteins become phosphorylation’s substrates at some point. This observation makes protein phosphorylation one of the most important PTM types, in addition to glucose and oxidative phosphorylation.
Protein regulation controls the protein’s function itself, its subcellular localization and its interaction with other proteins. Proteins being implicated in nearly every task of cellular life, their activation and deactivation is essential for the proper functioning of biological processes in living organisms.
Phosphopeptides synthesis service
SB-PEPTIDE can produce phosphopeptides with phosphorylation of serine, threonine and tyrosine and already successfully obtained peptides with 4 phosphorylations.
Thanks to Solid Phase Peptide Synthesis (SPPS), SB-PEPTIDE can incorporate one or more phosphorylated amino acids at precise positions during custom peptide synthesis. The peptide to be synthesized can contain non-phosphorylated serine/tyrosine/threonine and phosphorylated ones.
SB-PEPTIDE can produce series of peptides with phosphorylation at various positions to help finding the correct original position or to try everything possible when it is unknown.
Example:
Peptide 1 : XXX-Y-XX-Y-XXX-Y-XXX
Peptide 2: XXX-pY-XX-Y-XXX-Y-XXX
Peptide 3: XXX-Y-XX-pY-XXX-Y-XXX
Peptide 4: XXX-Y-XX-Y-XXX-pY-XXX
Phosphorylated peptides are compatible with other modifications such as biotinylation, incorporation of isotope labeled amino acids, conjugation to carrier proteins, etc.
Standard synthesis scales:

Contact us to get a quote.
Alternative stable phosphorylation
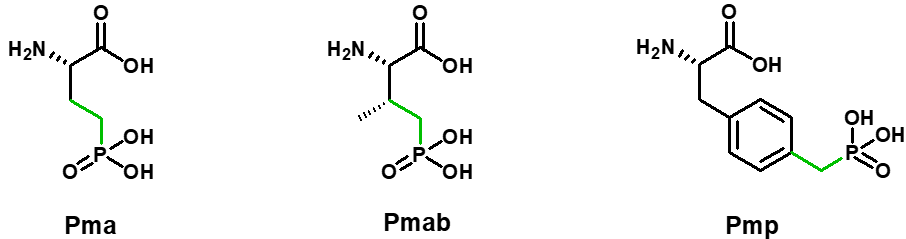
Over the past few years, alternative phosphorylated amino acids have been developed in order to maintain the in-vivo stability of phosphorylated peptides. Phosphorylated amino acids can be replaced by hydrolysis-stable amino acids functionalized with hydrolysis-stable phosphate groups Pma (Ser), Pmab (Thr) and Pmp (Tyr).
SB-PEPTIDE can produce peptides containing Pma, Pmab and Pmp. Contact us to get a quote.
References
Front. Genetics. 2014 Aug 07;5:270. doi: https://doi.org/10.3389/fgene.2014.00270
Physicochemical mechanisms of protein regulation by phosphorylation
Phosphorylation offers a dynamic way to regulate protein activity and subcellular localization, which is achieved through its reversibility and fast kinetics. Adding or removing a dianionic phosphate group somewhere on a protein often changes the protein’s structural properties, its stability and dynamics. Moreover, the majority of signaling pathways involve an extensive set of protein–protein interactions, and phosphorylation can be used to regulate and modulate protein–protein binding. Losses of phosphorylation sites, as a result of disease mutations, might disrupt protein binding and deregulate signal transduction. In this paper we focus on the effects of phosphorylation on protein stability, dynamics, and binding. We describe several physico-chemical mechanisms of protein regulation through phosphorylation and pay particular attention to phosphorylation in protein complexes and phosphorylation in the context of disorder–order and order–disorder transitions. Finally we assess the role of multiple phosphorylation sites in a protein molecule, their possible cooperativity and function.
MCP. 2013 Sep 13;12(12): 3453-3464. doi: https://doi.org/10.1074/mcp.R113.032862
The Coming of Age of Phosphoproteomics from Large Data Sets to Inference of Protein Functions
Protein phosphorylation is one of the most common post-translational modifications used in signal transduction to control cell growth, proliferation, and survival in response to both intracellular and extracellular stimuli. This modification is finely coordinated by a network of kinases and phosphatases that recognize unique sequence motifs and/or mediate their functions through scaffold and adaptor proteins. Detailed information on the nature of kinase substrates and site-specific phosphoregulation is required in order for one to better understand their pathophysiological roles. Recent advances in affinity chromatography and mass spectrometry (MS) sensitivity have enabled the large-scale identification and profiling of protein phosphorylation, but appropriate follow-up experiments are required in order to ascertain the functional significance of identified phosphorylation sites. In this review, we present meaningful technical details for MS-based phosphoproteomic analyses and describe important considerations for the selection of model systems and the functional characterization of identified phosphorylation sites.
Annu. Rev. Biomed. Eng. 2009 Aug 15;11:49-79. doi: https://doi.org/10.1146/annurev-bioeng-061008-124934
Proteomics by Mass Spectrometry: Approaches, Advances, and Applications
Mass spectrometry (MS) is the most comprehensive and versatile tool in large-scale proteomics. In this review, we dissect the overall framework of the MS experiment into its key components. We discuss the fundamentals of proteomic analyses as well as recent developments in the areas of separation methods, instrumentation, and overall experimental design. We highlight both the inherent strengths and limitations of protein MS and offer a rough guide for selecting an experimental design based on the goals of the analysis. We emphasize the versatility of the Orbitrap, a novel mass analyzer that features high resolution (up to 150,000), high mass accuracy (2–5 ppm), a mass-to-charge range of 6000, and a dynamic range greater than 103. High mass accuracy of the Orbitrap expands the arsenal of the data acquisition and analysis approaches compared with a low-resolution instrument. We discuss various chromatographic techniques, including multidimensional separation and ultra-performance liquid chromatography. Multidimensional protein identification technology (MudPIT) involves a continuum sample preparation, orthogonal separations, and MS and software solutions. We discuss several aspects of MudPIT applications to quantitative phosphoproteomics. MudPIT application to large-scale analysis of phosphoproteins includes (a) a fractionation procedure for motif-specific enrichment of phosphopeptides, (b) development of informatics tools for interrogation and validation of shotgun phosphopeptide data, and (c) in-depth data analysis for simultaneous determination of protein expression and phosphorylation levels, analog to western blot measurements. We illustrate MudPIT application to quantitative phosphoproteomics of the beta adrenergic pathway. We discuss several biological discoveries made via mass spectrometry pipelines with a focus on cell signaling proteomics.