MHC binding peptides prediction
Major Histocompatibility Complex (MHC),also known as the Human Leukocyte Antigens (HLA),is a group of genes that code for cells surface proteins. MHC molecules help the adaptive immune system to detect foreign substances by displaying endogenous or exogeneous antigens on their surface to enable their recognition by the appropriate T-cells.
MHC proteins types
Even though MHC is highly polygenic and polymorphic,the encoded molecules expressed on the cell’s surface can be divided in two subclasses known as MHC class I and MHC class II.
MHC class I
MHC class I proteins are expressed on all cells but red blood cells to present endogenous epitopes to Cytotoxic T-Cells (CTL or CD8+ T-cells). They are formed by one polypeptide chain constituted of three external domains (α1,α2,α3) and one constant region,called β2-microglobulin,presented on Figure 1 below.
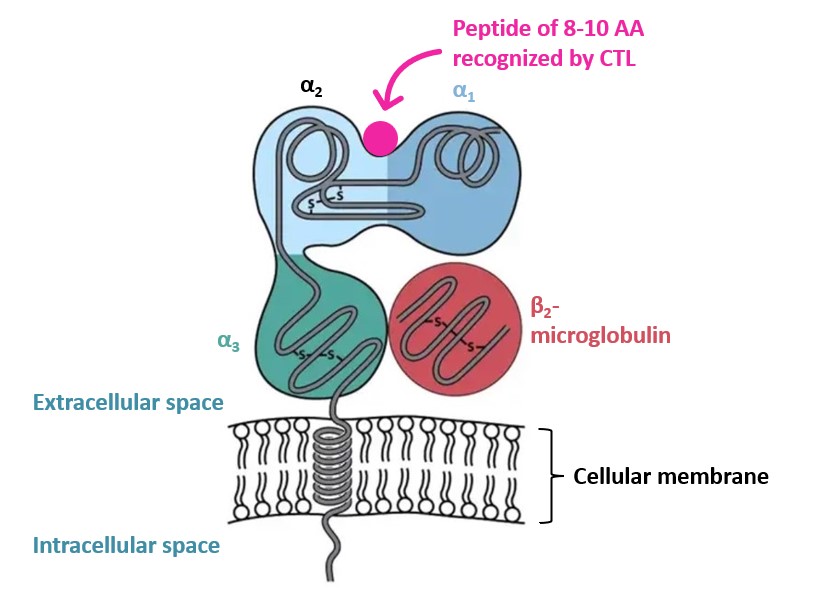
Figure 1 – MHC class I structure
MHC class II
On the other hand,MHC class II molecules are only expressed on Antigen Presenting Cells (APC),such as macrophages,B cells,and especially dendritic cells (DC). MHC class II proteins bind to exogeneous antigens to enable their detection by helper T-Cells (TH-cells or CD4+ T-cells). Class II MHC proteins structurally differs from the 1st class by being composed of two polypeptide chains constituted of two external domains each (α1,α2 and β1 , β2),depicted on Figure 2 below.
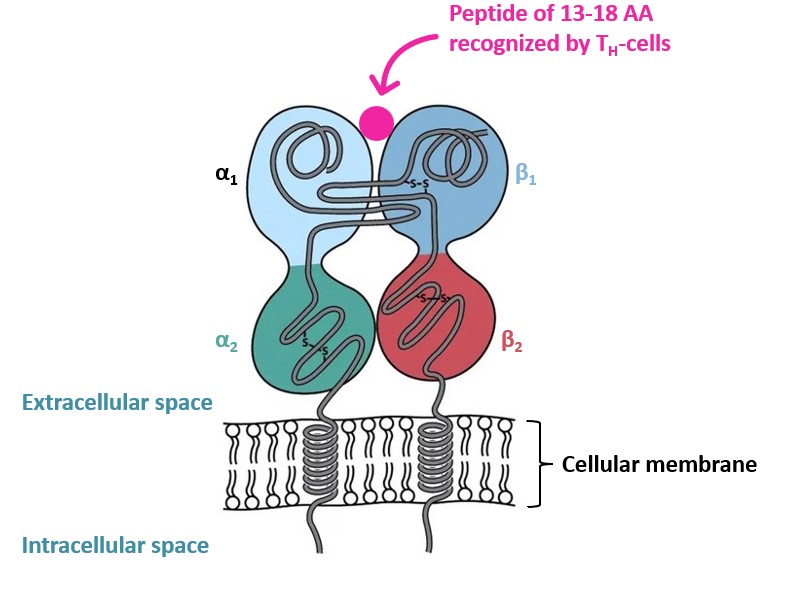
Figure 2 – MHC class II structure
Major Histocompatibility Complex and adaptive immunity
Unlike the innate immunity,the adaptive immune system is highly specific to each pathogen the body has been exposed to. This immunity strategy is involved in immunization modulation by creating immunological memory.
Among the cells involved in adaptive immunity processes,T-cells require MHC proteins to recognize pathogenic antigens. These immune cells display T-Cells Receptors (TCR) on their surface that is non-covalently associated with a complex of proteins known as CD3 and the ξξ homodimer.
The TCR complex (Figure 3 hereafter) is identical in all T-cells,but the co-receptors,thanks to which the appropriate T-cells recognize the corresponding MHC molecules,differ depending on the type of T-cells recruited i.e. CD4 or CD8 co-receptors for CTL or TH-cells,respectively.
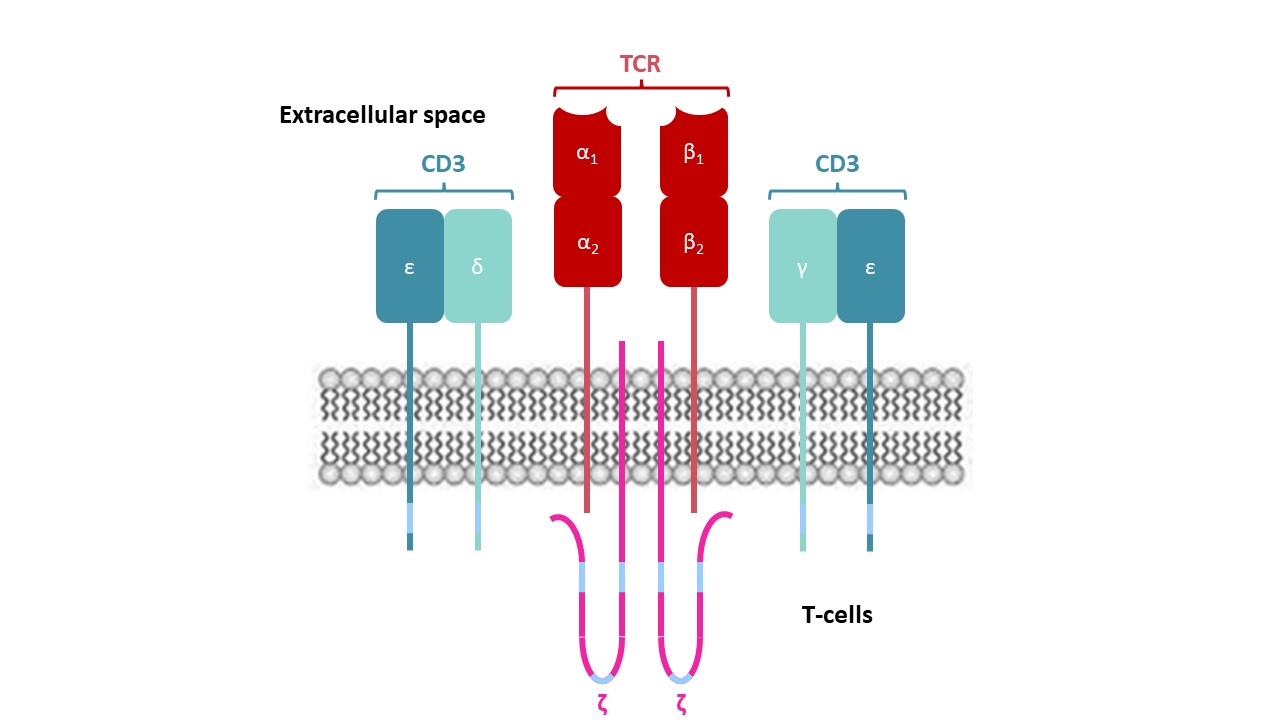
Figure 3 – TCR complex
Once the foreign epitope recognized by the corresponding T-cell,pre-programmed cell death via apoptosis will be initiated by CTL and naïve CD4+ T-cells will be activated in TH-cells that differentiate in memory or effector TH-cells to modulate immunization.
MHC binding peptides predictors
Nowadays,the specific activation of T-cells via MHC binding peptides is a highly valuable process to develop personalized cancer vaccines. Indeed,cancer cells express modified MHC molecules that are difficult to detect by the immune system. Fortunately,the synthesis of peptides able to bind these MHC proteins would enable the normal recognition and processing of tumor cells by immune cells.
To help design these synthetic peptides,various predictors were developed with free or paying access. Here is a non-exhaustive list of MHC peptide binding predictors:
IEDB – Immune Epitope Database
IEDB is a complete and powerful platform to identify MHC binding peptides,predict B-cell and T-cell epitopes and much more.
Discover IEDB platform
NetMHC: Binding of peptides to MHC class I molecules
NetMHC allows to predict allele-specific binding peptides to MHC class I.
Discover NetMHC platform
smm: Peptide-MHC Binding Prediction
Predicts peptide-MHC binding using the stabilized matrix method (SMM) algorithm with a specifically constructed amino acid substitution matrix
Discover smm MHC binding predictor
NetMHCpan
The NetMHCpan-4.1 server predicts binding of peptides to any MHC molecule of known sequence using artificial neural networks (ANNs).
Discover NetMHCpan
NetMHCcons
The NetMHCcons 1.1 server predicts binding of peptides to any known MHC class I molecule.
Discover NetMHCcons
SYFPEITHI
SYFPEITHI database comprises more than 7000 peptide sequences known to bind class I and class II MHC molecules.
Discover SYFPEITHI
Antijen
AntiJen v2.0,is a database containing quantitative binding data for peptides binding to MHC Ligand,TCR-MHC Complexes,T Cell Epitope,TAP,B Cell Epitope molecules and immunological Protein-Protein interactions. In addition,AntiJen has recently included Peptide Library,Copy Numbers and Diffusion Coefficient data.
Discover Antijen
MHCBN
The database provides information about allele specific MHC binding peptides,MHC non-binding,TAP binding,TAP non-binding peptides and T-cell epitopes reported in literature.
Discover MHCBN
Looking for a peptide provider ?
SB-PEPTIDE is a France-based company highly-specialized in peptides manufacturing and analysis. We offer a wide range of products and services dedicated to immunologists,including:
– Custom peptide synthesis: short to long peptides,cyclized,biotinylated,fluorescent,conjugated to KLH/BSA,…
– Synthesis of peptide libraries: peptide synthesis in 96 well-plates,fro 25€ / peptide
– Customized peptide pool: preparation of specific pools containing crude or purified peptides
– Catalog of antigens,epitopes,peptide pools (infection,cancer,control)
– Analytical services: SEC-HPLC MHC,mass spectrometry…
Feel free to browse our website and contact us for discussing your projects in more details.
Front. Immunol. 2017 Mar 17; 8. doi: https://doi.org/10.3389/fimmu.2017.00292
Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation
Antigen presentation by major histocompatibility complex (MHC) proteins is essential for adaptive immunity. Prior to presentation,peptides need to be generated from proteins that are either produced by the cell’s own translational machinery or that are funneled into the endo-lysosomal vesicular system. The prolonged interaction between a T cell receptor and specific pMHC complexes,after an extensive search process in secondary lymphatic organs,eventually triggers T cells to proliferate and to mount a specific cellular immune response. Once processed,the peptide repertoire presented by MHC proteins largely depends on structural features of the binding groove of each particular MHC allelic variant. Additionally,two peptide editors—tapasin for class I and HLA-DM for class II—contribute to the shaping of the presented peptidome by favoring the binding of high-affinity antigens. Although there is a vast amount of biochemical and structural information,the mechanism of the catalyzed peptide exchange for MHC class I and class II proteins still remains controversial,and it is not well understood why certain MHC allelic variants are more susceptible to peptide editing than others. Recent studies predict a high impact of protein intermediate states on MHC allele-specific peptide presentation,which implies a profound influence of MHC dynamics on the phenomenon of immunodominance and the development of autoimmune diseases. Here,we review the recent literature that describe MHC class I and II dynamics from a theoretical and experimental point of view and we highlight the similarities between MHC class I and class II dynamics despite the distinct functions they fulfill in adaptive immunity.
Nat Immunol. 2013 Oct;14(10):1014-1022. doi: https://doi.org/10.1038/ni.2703
Innate and adaptive immune cells in the tumor microenvironment
Most tumor cells express antigens that can mediate recognition by host CD8+ T cells. Cancers that are detected clinically must have evaded antitumor immune responses to grow progressively. Recent work has suggested two broad categories of tumor escape based on cellular and molecular characteristics of the tumor microenvironment. One major subset shows a T cell–inflamed phenotype consisting of infiltrating T cells,a broad chemokine profile and a type I interferon signature indicative of innate immune activation. These tumors appear to resist immune attack through the dominant inhibitory effects of immune system–suppressive pathways. The other major phenotype lacks this T cell–inflamed phenotype and appears to resist immune attack through immune system exclusion or ignorance. These two major phenotypes of tumor microenvironment may require distinct immunotherapeutic interventions for maximal therapeutic effect.
Cell. 2010 Mar 19;140(6):883-899. doi: https://doi.org/10.1016/j.cell.2010.01.025
Immunity,Inflammation,and Cancer
Inflammatory responses play decisive roles at different stages of tumor development,including initiation,promotion,malignant conversion,invasion,and metastasis. Inflammation also affects immune surveillance and responses to therapy. Immune cells that infiltrate tumors engage in an extensive and dynamic crosstalk with cancer cells,and some of the molecular events that mediate this dialog have been revealed. This review outlines the principal mechanisms that govern the effects of inflammation and immunity on tumor development and discusses attractive new targets for cancer therapy and prevention.
Catalog
Epitopes (OVA, MOG...)
T-helpers/adjuvants
Pools (CEF, PP65...)
Libraries (Spike...)
Peptide synthesis
mg to grs,crude to >98%
2 to +100 AA
10 days express service
Cyclization,biotin,dyes…
KLH/BSA conjugations
Peptide libraries & pools
Hundreds of peptides
Various scales available
Customized delivery format
Customized pools (matrix…)
Economical solutions
