Haemagglutinin (HA) peptide YPYDVPDYA – HA Tag
SB-PEPTIDE offers Human influenza hemagglutinin (HA) tag YPYDVPDYA,which is a well characterized 9 amino acids synthetic peptide deriving from an epitope of the original influenza hemagglutinin protein.
Haemagglutinin surface fusion glycoprotein for viral entry
Haemagglutinin (HA) (UniProt : P03437) is an envelope protein present in the viral membrane of influenza viruses. HA forms a trimer containing structurally distinct regions and an intact HA0 polypeptide composed of two subunits,called HA1 and HA2,connected by one disulfide bridge.
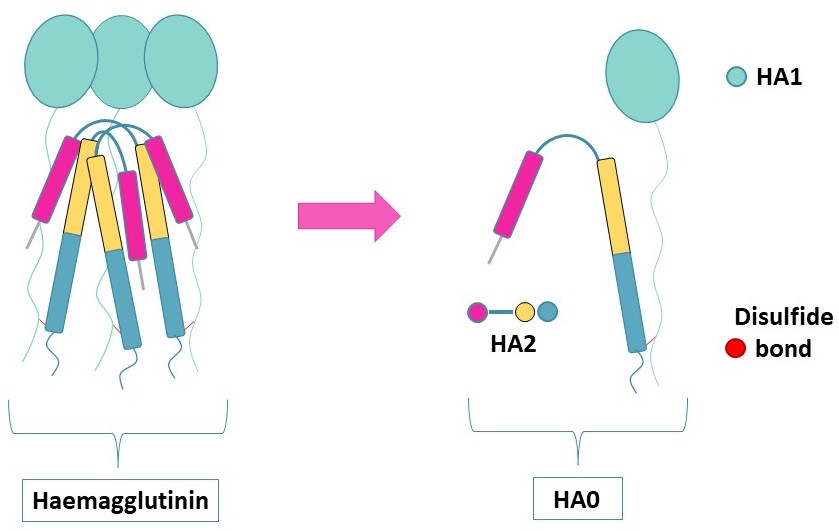
To initiate their fusion with their cellular hosts,influenza viruses first bind to sialic acids at the surface of respiratory epithelial cells. Following this interaction,they will enter the cell via endocytosis.
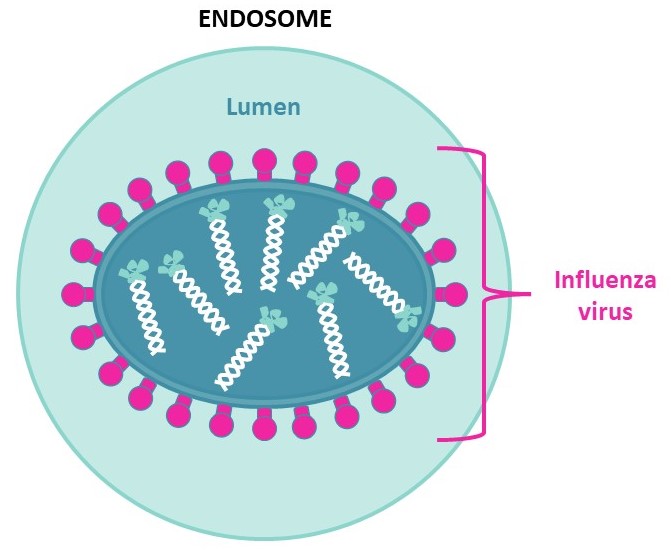
Due to proton pumps on the endosome’s membrane,the lumen will acidify under pH=6,cleaving HA1 from HA0 and thus freeing the N-terminal of HA2. This will allow HA2 anchoring to the host’s endosome,fusing the viral envelope to the endosome membrane to enable genome exchange and later viral multiplication.
Synthetic HA epitope – YPYDVPDYA [CAS: 92000-76-5]
HA synthetic peptide can be used for various applications. When coupled to a protein carrier,such as KLH,YPYDVPDYA is able to stimulate a host humoral immune response to generate monoclonal and polyclonal antibodies. In this case,the resulting antibodies are called « anti-tag antibodies »,whose aim is to optimize the detection and the purification of HA-tagged proteins.
More than the production of highly sensitive and specific anti-HA antibodies,YPYDVPDYA is used to compete with the latter to confirm binding specificity. This feature is also employed for HA-tagged protein purification to elute the protein of interest from affinity column of monoclonal anti-HA agarose.
SB-PEPTIDE is able to produce the biotinylated version of YPYDVPDYA peptide,as well as the KLH-conjugated version.
Visit our KLH peptide conjugation webpage here and do not hesitate to ask for a quote later on.
Technical specification
 |
Sequence : YPYDVPDYA |
 |
MW : 1102.15 g/mol (C53H67N9O17) |
 |
Purity : > 95% |
 |
Counter-Ion : TFA Salts (see option TFA removal) |
 |
Delivery format : Freeze dried in propylene 2mL microtubes |
 |
Other names : Anti-HA,HA1 fragment 98-106 |
 |
Peptide Solubility Guideline |
 |
Bulk peptide quantities available |
Price
| Product catalog | Size | Price € HT | Price $ USD |
| SB310-5MG | 5 mg | 250 | 315 |
| SB310-50MG | 50 mg | 1000 | 1250 |
References
Cell. 2016 May 12;165(7):1803-1817. doi: https://doi.org/10.1016/j.cell.2016.04.044
High-Throughput,High-Resolution Mapping of Protein Localization in Mammalian Brain by In Vivo Genome Editing
A scalable and high-throughput method to identify precise subcellular localization of endogenous proteins is essential for integrative understanding of a cell at the molecular level. Here,we developed a simple and generalizable technique to image endogenous proteins with high specificity,resolution,and contrast in single cells in mammalian brain tissue. The technique,single-cell labeling of endogenous proteins by clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9-mediated homology-directed repair (SLENDR),uses in vivo genome editing to insert a sequence encoding an epitope tag or a fluorescent protein to a gene of interest by CRISPR-Cas9-mediated homology-directed repair (HDR). Single-cell,HDR-mediated genome editing was achieved by delivering the editing machinery to dividing neuronal progenitors through in utero electroporation. We demonstrate that SLENDR allows rapid determination of the localization and dynamics of many endogenous proteins in various cell types,regions,and ages in the brain. Thus,SLENDR provides a high-throughput platform to map the subcellular localization of endogenous proteins with the resolution of micro- to nanometers in the brain.
Journal of Virology. 2015 Aug 12;89(20):10602-10611. doi: https://doi.org/10.1128/jvi.00939-15
Intermonomer Interactions in Hemagglutinin Subunits HA1 and HA2 Affecting Hemagglutinin Stability and Influenza Virus Infectivity
Influenza virus hemagglutinin (HA) mediates virus entry by binding to cell surface receptors and fusing the viral and endosomal membranes following uptake by endocytosis. The acidic environment of endosomes triggers a large-scale conformational change in the transmembrane subunit of HA (HA2) involving a loop (B loop)-to-helix transition,which releases the fusion peptide at the HA2 N terminus from an interior pocket within the HA trimer. Subsequent insertion of the fusion peptide into the endosomal membrane initiates fusion. The acid stability of HA is influenced by residues in the fusion peptide,fusion peptide pocket,coiled-coil regions of HA2,and interactions between the surface (HA1) and HA2 subunits,but details are not fully understood and vary among strains. Current evidence suggests that the HA from the circulating pandemic 2009 H1N1 influenza A virus [A(H1N1)pdm09] is less stable than the HAs from other seasonal influenza virus strains. Here we show that residue 205 in HA1 and residue 399 in the B loop of HA2 (residue 72,HA2 numbering) in different monomers of the trimeric A(H1N1)pdm09 HA are involved in functionally important intermolecular interactions and that a conserved histidine in this pair helps regulate HA stability. An arginine-lysine pair at this location destabilizes HA at acidic pH and mediates fusion at a higher pH,while a glutamate-lysine pair enhances HA stability and requires a lower pH to induce fusion. Our findings identify key residues in HA1 and HA2 that interact to help regulate H1N1 HA stability and virus infectivity.
Cell. 1998 Oct 01;95(3):409-417. doi: https://doi.org/10.1016/s0092-8674(00)81771-7
Structure of the Hemagglutinin Precursor Cleavage Site,a Determinant of Influenza Pathogenicity and the Origin of the Labile Conformation
The membrane fusion potential of influenza HA,like many viral membrane-fusion glycoproteins,is generated by proteolytic cleavage of a biosynthetic precursor. The three-dimensional structure of ectodomain of the precursor HA0 has been determined and compared with that of cleaved HA. The cleavage site is a prominent surface loop adjacent to a novel cavity; cleavage results in structural rearrangements in which the nonpolar amino acids near the new amino terminus bury ionizable residues in the cavity that are implicated in the low-pH-induced conformational change. Amino acid insertions at the cleavage site in HAs of virulent avian viruses and those of viruses isolated from the recent severe outbreak of influenza in humans in Hong Kong would extend this surface loop,facilitating intracellular cleavage.
Cell. 1982 Sep 01;30(2):649-656. doi: https://doi.org/10.1016/0092-8674(82)90261-6
Cell surface expression of the influenza virus hemagglutinin requires the hydrophobic carboxy-terminal sequences
We investigated the requirements of the carboxyterminal sequence for surface expression of the influenza viral hemagglutinin (HA). Deletions in the cloned hemagglutinin gene were introduced at locations upstream from and spanning into the region that codes for the hydrophobic carboxyl terminus. Primate cells infected with recombinants of the deleted HA gene and an SV40 vector were negative for surface immunofluorescence and failed to adsorb erythrocytes. Polypeptide analysis showed that the mutant hemagglutinins lacking the normal hydrophobic carboxy-terminal sequences were secreted into the medium. These data provide evidence that these sequences of the influenza hemagglutinin are responsible for accumulation at the cell surface. During infection with each deletion mutant,a truncated HA polypeptide was found intracellularly. Both intracellular and extracellular HAs were glycosylated,since a third species representing the unglycosylated mutant hemagglutinin was detected in the presence of tunicamycin. Interestingly,the secreted and intracellular mutant HA polypeptides differ from the surface HA in their sensitivity to endoglycosidase H,indicating that an alteration of glycosylation has occurred.
