SARS-CoV-2 Spike RBD 395-430 peptide
SARS-CoV-2 Spike RBD 395-430 peptide is an epitope of interest of the SARS-CoV-2 Spike S glycoprotein Receptor-Binding Domain (RBD). SARS-CoV-2 Spike RBD 395-430 peptide is useful for vaccine development and for structure-activity relationship studies.
SARS-CoV-2 Spike (S) glycoprotein
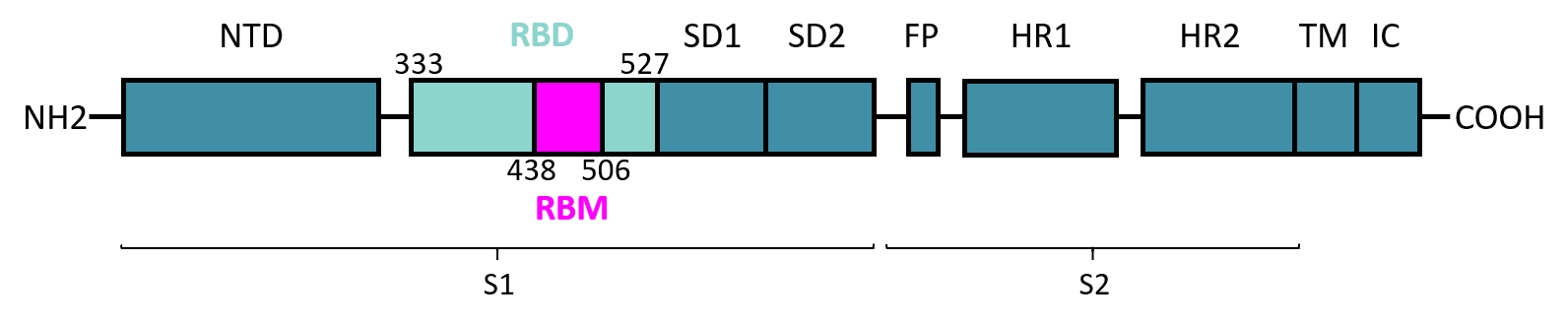
Spike (S) glycoprotein corresponds to one of the leading targets for COVID-19 disease. Present on the surface of Sars-CoV-2 virus,Spike S protein is a class I fusion protein that allows the virus to enter host cells.
With a 1 273 aa length,Spike protein has 2 subunits : S1 contains the receptor-binding domain RBD and S2 induces the fusion of the viral envelop with the cellular membrane.
SARS-CoV-2 Spike RBD
The receptor-binding domain in SARS-CoV-2 Spike protein allows binding to the Angiotensin-Converting Enzyme receptor 2 (ACE2 receptor) which mediates the viral entry.
SB-PEPTIDE also offers Biotin-SARS-CoV-2 Spike RBD 395-430 peptide.
Technical specification
 |
Sequence : VYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFT |
 |
MW : 4063 ,44 g/mol (C183H277N47O58) |
 |
Purity : > 95% |
 |
Counter-Ion : TFA Salts (see option TFA removal) |
 |
Delivery format : Freeze dried in propylene 2mL microtubes |
 |
Peptide Solubility Guideline |
 |
Bulk peptide quantities available |
Price
| Product catalog | Size | Price € HT | Price $ HT |
| SB091-1MG | 1 mg | 143 | 179 |
| SB091-5MG | 5 mg | 501 | 626 |
References
1- Jun Lan et al. Nature. 581(7807):215-220 (2020)

Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor
A new and highly pathogenic coronavirus (severe acute respiratory syndrome coronavirus-2,SARS-CoV-2) caused an outbreak in Wuhan city,Hubei province,China,starting from December 2019 that quickly spread nationwide and to other countries around the world1 ,2 ,3. Here,to better understand the initial step of infection at an atomic level,we determined the crystal structure of the receptor-binding domain (RBD) of the spike protein of SARS-CoV-2 bound to the cell receptor ACE2. The overall ACE2-binding mode of the SARS-CoV-2 RBD is nearly identical to that of the SARS-CoV RBD,which also uses ACE2 as the cell receptor4. Structural analysis identified residues in the SARS-CoV-2 RBD that are essential for ACE2 binding,the majority of which either are highly conserved or share similar side chain properties with those in the SARS-CoV RBD. Such similarity in structure and sequence strongly indicate convergent evolution between the SARS-CoV-2 and SARS-CoV RBDs for improved binding to ACE2,although SARS-CoV-2 does not cluster within SARS and SARS-related coronaviruses1 ,2 ,3 ,5. The epitopes of two SARS-CoV antibodies that target the RBD are also analysed for binding to the SARS-CoV-2 RBD,providing insights into the future identification of cross-reactive antibodies.
2- Wanbo Tai et al. Cell Mol Immunol. 17(6):613-620 (2020)

Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine
The outbreak of Coronavirus Disease 2019 (COVID-19) has posed a serious threat to global public health,calling for the development of safe and effective prophylactics and therapeutics against infection of its causative agent,severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2),also known as 2019 novel coronavirus (2019-nCoV). The CoV spike (S) protein plays the most important roles in viral attachment,fusion and entry,and serves as a target for development of antibodies,entry inhibitors and vaccines. Here,we identified the receptor-binding domain (RBD) in SARS-CoV-2 S protein and found that the RBD protein bound strongly to human and bat angiotensin-converting enzyme 2 (ACE2) receptors. SARS-CoV-2 RBD exhibited significantly higher binding affinity to ACE2 receptor than SARS-CoV RBD and could block the binding and,hence,attachment of SARS-CoV-2 RBD and SARS-CoV RBD to ACE2-expressing cells,thus inhibiting their infection to host cells. SARS-CoV RBD-specific antibodies could cross-react with SARS-CoV-2 RBD protein,and SARS-CoV RBD-induced antisera could cross-neutralize SARS-CoV-2,suggesting the potential to develop SARS-CoV RBD-based vaccines for prevention of SARS-CoV-2 and SARS-CoV infection.
- Amyloid Peptides
- Acetyl-ccbeta
- beta-Amyloid (1-11) Human
- beta-Amyloid (1-12) Human
- beta-Amyloid (1-14)
- beta-Amyloid (1-15) human
- beta-Amyloid (1-16) Human
- beta-Amyloid (1-17) Human
- beta-Amyloid (1-28) human
- beta-Amyloid (1-40) Human
- beta-Amyloid (1-6)-GGC Human
- beta-Amyloid (10-20) Human
- beta-Amyloid (11-20) Human
- Beta-Amyloid (12-20)
- beta-Amyloid (35-25)
- RNase A (77-82) Amyloidogenic peptide
- SEN 304
- Tau (195-209) Light
- Tau Peptide (45-73)
- Antimicrobial Peptides - AMP
- Abaecin
- Acetyl-Adhesin
- AcrAP1
- AcrAP1a
- AcrAP2
- AcrAP2a
- Alloferon 1
- Alloferon 2
- Alyteserin-1a
- Alyteserin-1b
- Alyteserin-1c
- Alyteserin-1d
- Alyteserin-2a
- Alyteserin-2b
- Alyteserin-2c
- Alyteserin-2Mb
- Anoplin
- Apidaecin IB
- B8R (20 – 27)
- Balteatide
- BMAP-18
- BMAP-18 (truncated)
- Buforin II
- Cecropin A
- Codesane
- CRAMP (1-39)
- CRAMP (6-39)
- CRAMP-18
- Cyclic L27-11
- Cycloviolacin O2 peptide
- Epinecidin-1
- FdM
- Feleucin-B01
- Gag (18-26) [Human immunodeficiency virus type 1] acetyl/amide
- Gag peptide [Simian immunodeficiency virus]
- Gag protein (181-189) acetyl/amide [Simian immunodeficiency virus]
- GLK-19
- hBD-2 peptide – human Beta Defensin 2
- HEL46-61
- Hepcidin-25 (LEAP-1 peptide)
- Histatin-5
- Histatin-8
- Hp1404
- Human Lysozyme (107-115)
- IDR-1
- Indolicidin
- Influenza Virus Nucleoprotein (311 – 325)
- KDAMP
- LL-13-37
- LL-17-29
- LL-17-32
- LL-37 amide
- LL-37 fragment (24-29)
- LL-37 fragment (30-34)
- LL-37 fragment (30-37)
- LL-37 peptide (CAP-18)
- Macropin 1
- Macropin 2
- Magainin II
- Magainin-1
- MALT1 substrate
- Mastoparan
- Mastoparan peptide
- mBD3 peptide – mouse Beta Defensin 3
- MP196
- N-formylated PSMalpha2
- N-formylated PSMalpha3
- P2-Hp-1935
- PA protein (Influenza A virus)
- PAF19
- PAF26
- Pantinin-1
- Pantinin-2
- Pantinin-3
- Pap12-6
- Parasin I
- Pseudin-2
- Sapecin B
- SARS-CoV-2 NSP13 (221-235)
- SARS-CoV-2 NSP13 (226-240)
- SARS-CoV-2 NSP13 (231-245)
- SARS-CoV-2 NSP13 (236-250)
- SARS-CoV-2 NSP13 (241-255)
- SARS-CoV-2 NSP13 (246-260)
- SARS-CoV-2 NSP13 (321-335)
- SARS-CoV-2 NSP13 (326-340)
- SARS-CoV-2 NSP13 (336-350)
- SARS-CoV-2 NSP13 (421-435)
- SARS-CoV-2 NSP13 (426-440)
- SARS-CoV-2 NSP13 (466-480)
- SARS-CoV-2 NSP13 (476-490)
- SARS-CoV-2 NSP13 (551-565)
- SARS-CoV-2 NSP13 (556-570)
- SARS-CoV-2 NSP13 (576-590)
- SARS-CoV-2 NSP13 (581-595)
- SARS-CoV-2 NSP7 (1-15)
- SARS-CoV-2 NSP7 (21-35)
- SARS-CoV-2 NSP7 (31-45)
- SARS-CoV-2 NSP7 (46-60)
- SARS-CoV-2 NSP7 (51-65)
- SARS-CoV-2 NSP7 (6-20)
- SARS-CoV-2 Nucleoprotein (1-17)
- SARS-CoV-2 Nucleoprotein (104-121)
- SARS-CoV-2 Nucleoprotein (126-140)
- SARS-CoV-2 Nucleoprotein (266-280)
- SARS-CoV-2 Nucleoprotein (271-285)
- SARS-CoV-2 Nucleoprotein (321-335)
- SARS-CoV-2 Nucleoprotein (331-345)
- SARS-CoV-2 Nucleoprotein (341-355)
- SARS-CoV-2 Nucleoprotein (346-360)
- SARS-CoV-2 Nucleoprotein (351-365)
- SARS-CoV-2 Nucleoprotein (353-370)
- SARS-CoV-2 Nucleoprotein (356-370)
- SARS-CoV-2 Nucleoprotein (51-65)
- SARS-CoV-2 Nucleoprotein (61-75)
- SARS-CoV-2 Nucleoprotein (86-100)
- SARS-CoV-2 Nucleoprotein 1 (56-70)
- SARS-CoV-2 Nucleoprotein 2 (326-340)
- SARS-CoV-2 ORF7a-10 (69-86)
- SARS-CoV-2 Spike (1192-1200)
- SARS-CoV-2 Spike (1196-1205)
- SARS-CoV-2 Spike (1197-1206)
- SARS-CoV-2 Spike (236-250)
- SARS-CoV-2 Spike (757-765)
- SARS-CoV-2 Spike (781-795)
- SARS-CoV-2 Spike (975-983)
- SARS-CoV-2 Spike (975-989)
- SARS-CoV-2 Spike (991-1000)
- SARS-CoV-2 Spike (996-1004)
- SARS-CoV-2 Spike (999-1007)
- SARS-inhibitor 4
- Scrambled LL-37 peptide
- Secretin (rat)
- Sendai Virus nucleoprotein (324-332)
- T22 peptide
- Temporin A
- Temporin L
- Tritrpticin
- VP4 (449-454) Nora virus
- VP4 (93-101) Nora virus
- [5-FAM]-LL-37
- [Cys]-Influenza Virus Nucleoprotein (311 – 325)
- Biotin Labeled
- BCL-6 corepressor Human (BCOR) (498-514) C-terminal Biotin
- beta-Amyloid (1-10) Biotin
- beta-Amyloid (1-11) Biotin
- beta-Amyloid (1-12) Biotin
- beta-Amyloid (1-13) Biotin
- beta-Amyloid (1-14) Biotin
- Biotin Apolipoprotein A-I (APOA1)(86-101)
- Biotin BRC4 (1517-1551)
- Biotin gliadin-derived peptide
- Biotin HER-2 substrate peptide
- Biotin phosphorylated CDK7 (157-169)
- Biotin phosphorylated JAK1 substrate peptide
- Biotin SBP1
- Biotin SBP2
- Biotin Steroid Receptor Coactivator-1 (SRC-1) (676-700)
- Biotin Substance P
- Biotin TAT (48-60)
- Biotin-aMp3
- biotin-aMptD
- Biotin-Axltide Peptide substrate
- Biotin-beta-Amyloid (1-15) human
- Biotin-Desmoglein-3 DSG3 (50-79)
- Biotin-GLP-1 (7-36)
- Biotin-Histone H3 (14-34) K23Me3
- Biotin-Histone H3 (14-34) pT22 K23Me3
- Biotin-Influenza A NP (147-155) (H-2Kd)
- Biotin-Jak2 substrate
- Biotin-LPETAG N-terminal Sortagging
- Biotin-LPETGG N-terminal Sortagging
- Biotin-Nrf2 (69-84)
- Biotin-PEG2-Claudin-3
- Biotin-PEG2-Claudin-6
- Biotin-PEG2-Claudin-9
- Biotin-TAT (47-57)
- Biotin-β Amyloid (1-42) Human
- Biotinylated L57
- C-Terminal Sortagging-AAA-[Lys(Biotin]
- C-Terminal Sortagging-[Lys(Biotin]
- Galanin (2-13)-Biotin
- Galanin (3-13)-Biotin
- Histone H2A (1-20)-GGK(Biotin)
- Histone H3 (1-20)
- Histone H3 (1-20)-[S]-Biotin
- Histone H3 (1-22) K4Me3-Biotin
- Histone H3 (1-22) K9Me1-Biotin
- Histone H3 (10-29)-Biotin
- Histone H3 (20-39)-Biotin
- MHC class II antigen E alpha (52-68)-Biotin
- Pyroglutamyl beta-Amyloid (4-14) Biotin
- [Biotin]-GLP-1
- Cancer Peptides
- A6 peptide
- AD01 N-terminal Q
- ARF peptide
- Bak BH3
- Bevacizumab Light chain
- Bid BH3 Peptide
- BIM 187
- Bim BH3, Peptide IV
- Braftide
- C7
- CooP
- Cyclo(CLLFVY)
- D-Arg PEP
- dodecapeptide AR71
- DOTA-(Tyr3)-octreotate Acetate Salt
- FAM49B (190-198) Mouse
- FFW
- FREG peptide
- GRGD-acid
- HIF-1 α (556-574)
- Infliximab Heavy chain (46-60)
- PEN-FFW
- Proapoptotic Peptide KLA
- RAGE antagonist peptide
- Rituximab Light chain (41-55)
- RKOpep
- XL 13m
- YSA acid
- [Tyr0]-Apelin-13
- Cell Penetrating Peptides - CPP
- (Arg)9 peptide
- (RFR)4XB
- 3xFlag [DYKDDDDK]
- Acetyl-Arg9
- Angiopep 2
- Antennapedia peptide
- Antennapedia peptide amide
- Antennapedia peptide Arg
- Azhx-Penetratin
- C105Y
- CPP9
- Cys(Npys)-Antennapedia peptide amide
- DYKDDDDK FLAG peptide
- Flexible Glycine Linker (3xGGGGS)
- L17E
- MART-1 (26-35)
- MHV EP™
- N3-His tag
- Penetratin peptide
- Pep – 1 – Chariot
- PR9
- RAG8
- Secretoneurin Mouse Rat
- T peptide
- TAT (47-57) peptide
- TfR targeting sequence
- [5-FAM]-(RXR)4XB
- Click Peptides
- COVID-19 Peptides
- ACE2 – Angiotensin-Converting enzyme 2 – peptide library
- Biotin-SARS-CoV-2 Spike RBD 319-335 peptide
- Biotin-SARS-CoV-2 Spike RBD 336-347 peptide
- Biotin-SARS-CoV-2 Spike RBD 348-357 peptide
- Biotin-SARS-CoV-2 Spike RBD 352-365 peptide
- Biotin-SARS-CoV-2 Spike RBD 371-394 peptide
- Biotin-SARS-CoV-2 Spike RBD 395-430 peptide
- Biotin-SARS-CoV-2 Spike RBD 513-520 peptide
- Biotin-SARS-CoV-2 Spike RBD 523-541 peptide
- Biotin-SARS-CoV-2 Spike RBM 438-458 peptide
- Biotin-SARS-CoV-2 Spike RBM 450-473 peptide
- Biotin-SARS-CoV-2 Spike RBM 480-496 peptide
- Biotin-SARS-CoV-2 Spike RBM 500-509 peptide
- CoV Main Protease (Mpro) Substrate
- SARS CoV-2 Spike (S) protein – Peptide library
- SARS-CoV 3C-like protease (3CLpro) substrate (C-terminal KK-acid)
- SARS-CoV-2 Membrane protein (141-158)
- SARS-CoV-2 Membrane protein (172-188)
- SARS-CoV-2 NSP7 (26-40)
- SARS-CoV-2 Nucleocapsid (N) protein – Peptide library
- SARS-CoV-2 Nucleoprotein 2 (261-275)
- SARS-CoV-2 ORF3a (26-40)
- SARS-CoV-2 Spike RBD 319-335 peptide
- SARS-CoV-2 Spike RBD 336-347 peptide
- SARS-CoV-2 Spike RBD 348-357 peptide
- SARS-CoV-2 Spike RBD 352-365 peptide
- SARS-CoV-2 Spike RBD 371-394 peptide
- SARS-CoV-2 Spike RBD 395-430 peptide
- SARS-CoV-2 Spike RBD 513-520 peptide
- SARS-CoV-2 Spike RBD 523-541 peptide
- SARS-CoV-2 Spike RBM 438-458 peptide
- SARS-CoV-2 Spike RBM 450-473 peptide
- SARS-CoV-2 Spike RBM 480-496 peptide
- SARS-CoV-2 Spike RBM 500-509 peptide
- Spike Protein (SARS-CoV-2) Peptide Pool
- UCI-1
- Variants package of SARS CoV-2 Spike (S) protein mutation – Peptide library
- Fluorescent Peptides
- (Cbz-LGR)2-[Rh110]
- (N-Cbz-Nle-KRR)2-[Rh110]
- (PFR)2-[Rh110]
- (Tos-GFHR)2-[Rh110]
- (Tos-GPR)2-[Rh110]
- (Tos-YASR)2-[Rh110]
- 5-FAM-Fz7-21
- AAA-C(AF647) C-Terminal Sortagging
- Ac-Arg-Gly-Lys(Ac)-AMC
- Ac-GPLD-[Rh110]-[D-Pro]
- Ac-RGK-[AMC]
- Ac-RLR-[AMC] Proteasome Substrate
- Ac-RLR-[Rh110]-[D-Pro]
- Acetyl-Alpha-2-antiplasmin-[AF680]
- Acetyl-Histone H4 (1-21) K5Ac, K8Ac, K12Ac, K16Ac-GG-[Lys(5-FAM)]
- Acetyl-Histone H4 (1-23) K16Ac-GG-[Lys(5-FAM)]
- Acetyl-Histone H4 (1-23)-GG-[Lys(5-FAM)]
- acfTAT
- ACTH (1-24) -[5-FAM]
- AF488 6xHis Tag
- AF488 Insulin
- AF488 Plectin-1-targeting peptide
- AF647 RGD peptide
- C-terminal Sortagging-[Cys(AF488)]
- C-terminal Sortagging-[Cys(AF680)] acid
- C-terminal Sortagging-[Cys(AF680)] amide
- C-terminal Sortagging-[Cys(Sulfocyanine3)]
- C-terminal Sortagging-[Cys(Sulfocyanine5)]
- C-terminal Sortagging-[Cys(Sulfocyanine7)]
- Cathepsin G FRET substrate [5-FAM]/[6-TAMRA]
- Cys(BDP630/650)-Galanin (1-30) Human
- DNA damage-binding protein 2 (DDB2)-[Cys(AF647)]-amide
- ERAAP substrate Ep
- Exendin-4 [Lys(AF647)]
- FLAG tag (Cy3B)
- Fluorescein HLA-A*02:01 HBV core (18-27)
- Formyl-MLF-[Cys(AF488)]
- GGG-C(AF647) C-Terminal Sortagging
- GGG-[K(5-TAMRA)] C-terminal Sortagging
- Ghrelin-[Cys(AF647)] Human
- GLP-1 (7-36) [Cys(Sulfocyanine5)]
- Glucagon (1-29)-[Cys(Cy5)]
- Glucagon (1-29)-[Lys(AF647)]
- GRGD-[Cys(AF647)]
- H-Met-Gly-Pro-[AMC].HCl
- HCV NS3 protease FRET substrate
- Histone H3 (1-15) K4Me3, K9Ac, pS10
- Histone H3 (1-20) K4Me2-GG-[Lys(5-FAM)]
- Histone H3 (1-20) K4Me3, K9Ac, pS10-GG-[Cys(Aurora™ Fluor 647)]
- Histone H3 (1-20) K4Me3, K9Ac, pS10-GG-[Lys(5-FAM)]
- Histone H3 (1-20) K4Me3, K9Ac-GG-[Lys(5-FAM)]
- Histone H3 (1-20) K4Me3, pS10-GG-[Lys(5-FAM)]
- Histone H3 (1-20) K4Me3-GG-[Cys(Aurora™ Fluor 647)]
- Histone H3 (1-20) K4Me3-GG-[Lys(5-FAM)]
- Histone H3 (1-20) pT3, K4Me3-GG-[Lys(5-FAM)]
- Histone H3 (1-20)-GG-[Cys(Aurora™ Fluor 647)]
- Histone H3 (1-20)-GG-[Lys(5-FAM)]
- Histone H3-GG-[Cys(5-FAM)]-amide
- Insulysin FRET substrate [Mca]/[Dnp]
- KHLF-[AMC]
- LasB FRET substrate
- Leu-AFC.HCl
- Melittin [Cy5]
- PFR-[AMC]
- Plasmin-[AMC] substrate
- Ser3-n-octanoyl Ghrelin-[Cys(AF647)] Human
- SMAC/DIABLO [Lys(5-FAM)]
- SMAC/DIABLO-[Cys(AF647)]
- SMRT peptide-(Cys[AF633])
- Suc-LLVY-[AMC]
- Suc-LLVY-[Rh110]-[D-Pro]
- [β-Ala]-[Lys(5-TAMRA)]-acid
- [β-Ala]-[Lys(AMCA)]-acid
- [5-FAM] Antennapedia peptide amide
- [5-FAM] EGFR/kinKDR peptide substrate
- [5-FAM] Histone H3 (1-14) K4Me3
- [5-FAM] Kemptide
- [5-FAM]-(KFF)3K
- [5-FAM]-(RFR)4XB
- [5-FAM]-Arg9
- [5-FAM]-beta-Amyloid (1-15) Human
- [5-FAM]-C7
- [5-FAM]-CADY
- [5-FAM]-Collagen alpha-1(I)-(5-TAMRA)
- [5-FAM]-CRAMP (6-39)
- [5-FAM]-EB1
- [5-FAM]-ERKtide
- [5-FAM]-Galanin (1-30) Human
- [5-FAM]-GLP-1
- [5-FAM]-GLP-1 (7-36)
- [5-FAM]-IFN-γ receptor (pTyr) peptide
- [5-FAM]-M918
- [5-FAM]-MAP
- [5-FAM]-MPG∆NLS
- [5-FAM]-PR9
- [5-FAM]-PTH (1-34)
- [5-FAM]-pVec
- [5-FAM]-RGD peptide
- [5-FAM]-RKOpep
- [5-FAM]-RPKPQQFFGLM-NH2
- [5-FAM]-SRC Substrate Peptide
- [5-FAM]-TAT
- [5-FAM]-TAT (47-57) amide
- [5-FAM]-Tp10
- [5-FAM]-Tyr-Ahx-Ser-Asp-Lys-Pro-acid
- [5-FAM]-Val
- [5-FAM]-VGB4
- [5-FAM]/[Lys(Dabcyl)]-CoV Main Protease (Mpro) Substrate
- [5-FAM]/[Lys(Dnp)]-SARS-CoV-2 S1/S2
- [5-TAMRA] Galanin, Human
- [5-TAMRA]-ATIA agonist [sar1, Ile4, Ile8]
- [5-TAMRA]-Galanin (1-30) Human
- [5-TAMRA]-LPETAG N-terminal Sortagging
- [5-TAMRA]-LPETGG N-terminal Sortagging
- [5-TAMRA]/[Lys(BHQ-2)] Ubiquitin
- [5-TAMRA]/[Lys(BHQ-2)]-CoV Main Protease (Mpro) Substrate
- [6-FAM]-Arg8
- [Atto655]-LifeAct (Abp140 1-17)
- [Aurora™ Fluor 647]-RGD peptide
- [Azhx]-[Lys(Mca)]-P11-8
- [BDP630/650]3-halphaCGRP (calcitonin gene-related peptide)
- [Cy3B]-LifeAct (Abp140 1-17)
- [Cys(AF488)]-Penetratin
- [Cys(AF647)]-Jak2/3 substrate
- [DABCYL]/[Glu(EDANS)] SARS-CoV-2 3C-like protease (3CLpro) substrate
- [FITC]-Ahx-(KKEEE)3K carrier peptide
- [FITC]-C7
- [FITC]-pCREB (127-134) substrate
- [MCA]/[Lys(Dnp)]-CoV Main Protease (Mpro) Substrate
- [Rhodamine Green]-LifeAct (Abp140 1-17)
- [Sulfo-Cyanine3]-LifeAct (Abp140 1-17)
- [Sulfo-Cyanine5]-Val-Pro-Valp(OPh)2
- [TAMRA]-beta-Amyloid (1-15) Human
- ™PRSS4 (199-207) fluorogenic peptide
- GPCR Modulators
- Growth Factors and Cytokines
- Boc-Val-Pro-Arg-AMC
- CHKtide
- CSK substrate
- EGFR (963-975)
- EGFR/kinKDR peptide substrate
- HSP70/DnaK Substrate Peptide
- IRS-1 substrate
- LHRH
- N-methylated ERAP1substrate
- Phosphorylated CHKtide
- Phosphorylated Sakamototide
- PKA Substrate
- Renin substrate
- Sakamototide
- SAMS peptide
- Somatostatin 14 (human, rat, mouse, pig, chicken, frog)
- SRC Substrate Peptide
- Hematology Related Peptides
- Histone Peptides
- Acetyl-Histone H4 (1-21)
- H4 peptide (16-23)
- Histone H1 derived peptide
- Histone H2A (1-20)
- Histone H2A (78-86)
- Histone H3 (1-18)
- Histone H3 (1-20) K4Me3
- Histone H3 (1-20) K4Me3, K9Ac, pS10-GG-Biotin
- Histone H3 (1-21)
- Histone H3 (1-21) K4ac
- Histone H3 (1-21) K4Me2
- Histone H3 (1-21) K4Me3
- Histone H3 (1-21) K9Me2
- Histone H3 (1-22)
- Histone H3 (1-8)
- Histone H3 (20-36) K27Me3
- Histone H3 (22-30) K27Me3
- Histone H3 (30-41) K36Me2
- Histone H3 (32-38) K36Me2
- Histone H3 (32-47)
- Histone H3-GGC-amide
- Histone H3.2 (1-44)
- Histone H3.3 (1-44)
- Histone H4 (1-21)
- Histone H4 (1-21) R3Me1
- Histone H4 (1-21) R3Me2
- Histone H4 (1-23)
- Osteogenic Growth Peptide (OGP)
- SETD8 Peptide
- Hormones
- ACTH (7-39) human
- Alexamorelin
- Alpha mating factor – WHWLQLKPGQPMY
- ANP (1-23)
- ANP (13-26)
- ANP (7-20)
- ANP (7-23)
- ANP (9-22)
- ANP 1-28 Human
- BNP-32 human
- Bradykinin
- Calcitonin, human – Agonist of the calcitonin receptor CTR
- Calcitonin, Rat
- Calcitonin, Salmon
- CCL2 (MCP-1)
- Exendin 3 (9-39) amide
- Exendin 4 (4-39)
- Exendin 4 – Potent GLP-1R agonist
- Gastrin Releasing Peptide, human
- GIP (1-42)-[C] human
- GIP (Pro 3)
- GIP, human
- GLP-1 (1-37)
- GLP-1 (7-36) amide
- GLP-1 (9-36) amide
- Glucagon (3-29)
- Glucagon like-peptide-2 (GLP-2)
- GPS1573
- GRP (14-27), human, porcine
- h-Chemerin-9 (149-157)
- Insulin beta Chain Peptide (15 – 23)
- Isotocin
- Liraglutide
- Motilin (1-10)
- Motilin (1-12)
- Motilin (human, porcine)
- Oxytocin
- Pro-BNP (47-76)
- Protirelin – Thyrotropin-releasing hormone (TRH) (CAS: 24305-27-9)
- PTH (1-13) Human
- PTH (1-34) human
- PTH (13-34) Human
- RANTES (CCL5)
- [Glu2]-TRH peptide (CAS: 85541-78-2)
- Immunology - Antigens/Epitotes/Pools/Librairies
- 2-Furoyl-LIGRL-amide
- 4-Fluorobenzoyl-A20FMDV2
- A*02:01/Human Survivin (96-104) peptide – LTLGEFLKL
- A*02:01/Human Survivin/SurA2.M (LMLGEFLKL) peptide
- AAV8 capsid protein
- Acetyl-HIV-1 reverse transcriptase (A2-YI9)
- Acetyl-TGF-beta 2-LAPbeta (259- 269)
- AF12198
- AH1 Sequence (6-14)
- AIP-I
- AIP-II
- AIP-III
- AIP-IV
- Allergen Ara h 1 (560-572)
- alpha-Gliadin (31 – 43)
- Annexin A1 (2-12)
- Ara h 2 (147-155) peanut Allergen
- Ara h 3 (278-284) peanut Allergen
- Ara h 6 (120-131) peanut Allergen
- Ara h1 (555-577) peanut Allergen
- AYPGFK Protease-Activated Receptor-4 (PAR-4)
- B-peptide
- BAM (8-22)
- BAT3 (340-347), human
- BDC2.5 mimotope 1040-51
- Biotin-FluM1 (58-66) peptide
- Biotin-MAGE-A1 (278-286) peptide
- Biotin-MAGE-A2 (157-166) peptide
- Biotin-NY-ESO-1 (157-165) C165V peptide
- Biotin-Ova (323-339) peptide
- Biotin-PADRE peptide AKFVAAWTLKAAA
- C5A
- CD20 (188-196) peptide – SLFLGILSV
- CEF (HLA Class I Control) Peptide Pool
- CEFT Control Peptide Pool (HLA Class I Control)
- Cilengitide (Linear)
- CMV IE-1 (213-225)
- CMV pp65 (113-121) peptide – VYALPLKML
- CMV pp65 (120-129) peptide – MLNIPSINV
- CMV pp65 (415-429) (HLA-B7)
- CMV pp65 (485-500)
- CMV pp65 (495-503) (HLA-A2)
- CMV pp65 (511-525)(HLA-B44)
- Compstatin
- Cyclo(-RGDyK)
- Cyclo(RGDfK)
- EBV BMLF1 (280-288) (HLA-A2)
- EBV BMLF1 (280-288) peptide – GLCTLVAML
- EBV BNRF1 (1238-1252)
- EBV BRLF1 (134-142) (HLA-A11)
- EBV BRLF1 (148-156) (HLA-A3)
- EBV BRLF1 (28-37) (HLA-A24)
- EBV BZLF1 (190-197) (HLA-B8)
- EBV BZLF1 (40-48) (HLA-E)
- EBV EBNA3A (158-166) (HLA-B8)
- EBV EBNA3A (325-333) (HLA-B8)
- EBV EBNA3A (379-387) (HLA-B7)
- EBV EBNA3A (458-466) (HLA-B35)
- EBV EBNA3A (603-611) (HLA-A3)
- EBV EBNA3B (416-424) (HLA-A11)
- EBV EBNA3C (258-266) (HLA-B27)
- EBV EBNA3C (281-290) (HLA-B44)
- ELA Elabela/Toddler-32
- Fibrinogen, b43-63
- Fibrinopeptide
- Flu-HA-B (306-318) MHC II DRB1*01:01
- FluM1 (58-66) peptide – GILGFVFTL
- GHK tripeptide
- gp100 (25-33) epitope – KVPRNQDWL
- gp100 (280-288) A288V HLA-A*0201 antigen peptide – YLEPGPVTV
- GP33 (1-9)
- gp96-II
- Haemagglutinin (HA) peptide YPYDVPDYA
- HCV NS5B (2594-2602) peptide – ALYDVVTKL
- Her-2/neu (85-94) peptide – LIAHNQVRQV
- HIV-1 p17 Gag (77-85) peptide – SLYNTVATL
- HIV-1 reverse transcriptase (A2-YI9)
- HLA-A*02:01 HBV core (18-27)
- HLA-A*02:01 NY-ESO-1 (157-165)
- HLA-A*02:01 Polymerase (400-408)
- HLA-A*02:01 Polymerase (417-425)
- HLA-DRB1*1501 peptide
- HPV E7 protein (49-57)
- HPV16 E7 (86-93)
- HS1 protein (160-168)
- hsBCL9CT-24
- Human PD – L1 inhibitor V
- I-A(g7) BDC2.5 mimotope
- IDR 1002
- IFNB1 (118-132) Human
- IL-33 peptide
- Influenza A HA (306-318)
- Influenza A NP (265-273) (HLA-A3)
- Influenza A NP (380-388) (HLA-B8)
- Influenza A NP (383-391) (HLA-B27)
- Influenza A NP (44-52) (HLA-A1)
- Influenza A NP (91-99) (HLA-A68)
- Influenza A PB1 (591-599) (HLA-A1)
- Interleukin-27 subunit beta (22-30)
- KD20 peptide
- MAGE-A p248V9 peptide – YLEYRQVPV
- MAGE-A p248V9 scrambled (RQYVELPYV)
- MAGE-A1 (278-286) peptide – KVLEYVIKV
- MAGE-A1 (278-286) scrambled (ELIVKVYKV)
- MAGE-A2 (157-166) peptide – YLQLVFGIEV
- MAGE-A2 (157-166) scrambled (VLVYFQEIGL)
- MAGE-A3 (112-120) peptide – KVAELVHFL
- MAGE-A3 (FLWGPRALV)
- MAGE-A3 (IMPKAGLLI)
- MBP-B MHC II DRB1*15:01 (84-102)
- Melan-A (26-35) A27L peptide – ELAGIGILTV
- Melan-A (26-35) peptide – EAAGIGILTV – CAS: 156251-01-3
- Melan-A (26-35) scrambled (AIEIAGGLTV)
- MHC binding peptides prediction
- MOG (34-56) Human amide
- MOG (35-51) cit46 human
- MOG (35-55) amide Mouse, Rat
- MUC1 (12-20) peptide – LLLLTVLTV
- Myoglobin 137-148 MHC II DRB1*03:01
- Nangibotide
- NY-ESO-1 (123-137) DRB1*04:01 peptide – LKEFTVSGNILTIRL
- NY-ESO-1 (157-165) C165V peptide – SLLMWITQV
- NY-ESO-1 (157-165) C165V scrambled (MSILWQLVT)
- NY-ESO-1 (157-165) peptide – SLLMWITQC
- ORF65 (131-140) [Murid herpesvirus 4]
- OVA (250-264)
- OVA (251-264)
- OVA (323 – 339) amide
- Ova (323-339) peptide – ISQAVHAAHAEINEAGR
- OVA 257 264 peptide – SIINFEKL
- OVA 257 264 peptide SIINFEKL – KLH conjugate
- OVA 257-264 scrambled (FILKSINE)
- OVA Peptides Pool
- Ovalbumin (324-338), chicken, quail
- Ovalbumin (324-340) acetyl/amide, chicken
- ovalbumin (371-382), chicken
- P17
- PADRE peptide – AKFVAAWTLKAAA
- Palmitoyl GHK tripeptide
- PD-1 (21-35)
- PD-1 (24-38)
- PD-1 (27-41)
- Peptide antigen and epitope catalog
- Peptide Tyrosinase (Asp371) – HLA-A*0201 (YMDGTMSQV)
- Pip6a
- PLP (139-151)
- PMX 205
- PMX 53
- PRAME (100-108) HLA-A*0201
- Restricted PADRE peptide- ak(Cha)VAAWTLKAAa-Ahx-C
- RS09/Toll Like Receptor TLR4 agonist – APPHALS – CAS 1449566-36-2
- S2-16
- Se™elanotide
- SMAP-18
- SPA4 Peptide
- survivin (baculoviral IAP repeat-containing protein 5) (21-28)
- SYFPEITHI – MHC binding peptide
- TET 830 modified/T-helper epitope from tetanus toxoid – AQYIKANSKFIGITEL
- TET 830/Tetanus Toxin (830-844) peptide – QYIKANSKFIGITEL
- Tetanus Toxin P2 (830 – 844)
- TetTox-B (830-843) MHC II DRB1*07:01
- TKD (450-463)
- Tregitope 084
- Tregitope 289
- TRP-2 Peptide (180-188) – SVYDFFVWL
- Uty HY Peptide (246-254) Mouse
- V5 peptide
- Vitronectin (367-378)
- [Ala144]-PLP (139-151)
- Ion Channels and Transporters
- (Dap22)-ShK
- 8xHis-ProTx-II
- Aah-II
- Acetyl-Claudin-3
- Acetyl-Claudin-6
- Acetyl-Claudin-9
- ACT1
- ADWX-1
- Agitoxin-2
- alpha-cobratoxin
- AmmTx3
- Apamin
- APETx2
- Apolipoprotein A-I (APOA1)(86-101)
- Apolipoprotein KV domain (67 – 77)
- ATTO488-Charybdotoxin
- ATTO488-ProTx-I
- ATTO488-ProTx-II
- ATX-II
- BDS-I
- BeKm-1
- Biotin-ProTx-I
- BmP02
- C-Peptide (57-87) human
- Charybdotoxin
- Chlorotoxin
- Conantokin-G
- Crotamine
- Cy3-Crotamine
- Cy5-Huwentoxin-IV
- Cy5-ProTx-I
- Cy5-ProTx-II
- D-GsMTx4
- Dc1a
- Echistatin α1 isoform
- GAP26
- GaTx2
- GrTx1
- GsAF-1
- GsAF-2
- GsMTx4
- Guangxitoxin-1E
- Hainantoxin-III
- Hainantoxin-IV
- Hm1a
- HSA (55-66)
- HsTx1
- Huwentoxin-I
- Huwentoxin-IV
- Huwentoxin-XVI
- Iberiotoxin
- Jingzhaotoxin-34
- Jingzhaotoxin-III
- Kaliotoxin-1
- Latartoxin-1a
- Leiurotoxin-1
- Leiurotoxin-1 Dab7
- Lys-conopressin-G
- Mambalgin-1
- Margatoxin
- Maurocalcine
- Maurotoxin
- Melittin
- MitTx (MitTx-α + MitTx-β)
- Morphiceptin
- MT7 – Muscarinic Toxin 7
- NMB-1
- Obtustatin
- OD1
- Panx-1 mimetic inhibitory peptide
- Phlotoxin-1
- Phrixotoxin-2
- Phrixotoxin-3
- PNC 27
- ProTx-I
- ProTx-II
- ProTx-II-Biotin
- ProTx-III
- Psalmotoxin-1 (PcTx1)
- Purotoxin-1
- Rho-Conotoxin-TIA
- ShK – Stichodactyla toxin
- Stromatoxin-1 (ScTx1)
- Tamapin
- TAMRA-Charybdotoxin
- TAMRA-ShK
- Tertiapin Q
- Tf2 scorpion toxin
- U2-sicaritoxin-Li1a
- Waglerin-1
- Waglerin-1-FAM
- α-Conotoxin BuIA
- α-Conotoxin PIA
- α-conotoxin-GI
- α-conotoxin-GID
- α-conotoxin-IMI
- α-conotoxin-MI
- α-conotoxin-PeIA
- αC-Conotoxin-PrXA
- β-Pompilidotoxin
- µ-conotoxin KIIIA
- µ-conotoxin-CnIIIC
- µ-conotoxin-GIIIB
- μ-conotoxin-PIIIA
- µO conotoxin MrVIB
- ρ-Da1a (AdTx1)
- ω-agatoxin-IVA
- ω-Conotoxin-GVIA
- ω-Conotoxin-MVIIA
- ω-Conotoxin-MVIIC
- ω-Conotoxin-SO3
- ω-Hexatoxin-Hv1a
- ω-Tbo-IT1
- Multiple sclerosis peptides
- Biotin-Mouse MOG (35-55) peptide
- Experimental Autoimmune Encephalomyelitis KIT
- Human MOG Peptides Pool
- MBP (1-11) human: Ac-ASQKRPSQRHG CAS 106128-98-7
- MBP (84-97) – VVHFFKNIVTPRTP
- MBP (85–99) – EKPKVEAYKAAAAPA
- MOG (183-191) – FVIVPVLGP
- MOG (35-55), human peptide
- MOG (91-108) peptide – SDEGGYTCFFRDHSYQEE
- MOG (92-106) peptide – DEGGYTCFFRDHSYQ
- MOG (97-108) peptide- TCFFRDHSYQEE
- Mouse MOG (35-55) peptide
- PLP (139-151) peptide – HSLGKWLGHPDKF – [CAS 122018-58-0]
- PLP (178-191): NTWTTCQSIAFPSK mouse, rat
- Myelin Basic Protein (MBP) Peptides
- Neuroscience Peptides
- (Ala11, D-Leu15)-Orexin B human
- (Arg8) Vasopressin (AVP)
- (Arg8) Vasotocin
- (D-Pro7)-Angiotensin I/II (1-7)
- (Des-octanoyl)-Ghrelin Human
- Acein
- Acetyl-Alpha-synuclein (1-13)
- Acetylated alpha-synuclein (1-7) amide
- ACTH (1-10) Human
- ACTH (1-17) Human
- ACTH (1-24) Human
- ACTH (1-39) Human
- ACTH (11-24)
- ACTH (15-24) Cys
- ACTH (18-39) Human
- ACTH (7-38) Human
- ACTH (7-39) Cys
- Alpha-Casozepine
- alpha-MSH
- Alpha-synuclein (1-13)
- Amylin (1-37) Human
- Amyloid beta peptides
- Angiotensin (Human, 1-7)
- Angiotensin I
- Angiotensin II Antipeptide
- Angiotensin III
- Angiotensin IV (3-8)
- Apelin (65-76), human
- Apelin-17 (human, bovine)
- Apolipoprotein E fragment (133-149) – COG133 : LRVRLASHLRKLRKRLL (CAS : 514200-66-9)
- Biotin-ACTH (1-39) Human
- CCK octapeptide Cholecystokinin (26-33)
- CE dipeptide
- CMX-8933
- CRF human, rat
- Duck liver-derived peptide 2
- Duck liver-derived peptide 3
- Duck liver-derived peptide 4
- EC dipeptide
- Echinotocin neuropeptide
- FMRFamide peptide
- Galanin (1-13)
- Galanin (1-15) Porcine, Rat
- Galanin (1-17) Porcine
- Galanin (13-20) Mouse
- Galanin (2-12) acid
- Galanin (2-13)
- Galanin (2-13) acid
- Galanin (2-30) acid
- Galanin (3-13)
- Galanin Human
- Galanin Mouse, Rat
- Galantide
- Ghrelin Human
- Ghrelin Rat, Mouse
- GP dipeptide
- GRP (18-27) (human, porcine, canine)
- Hyp-Gly dipeptide
- IFNB1 (118-132) Human deimmunised
- Kinetensin
- Kisspeptin 10 human
- Kisspeptin 14 human
- KLPGF peptide
- L57
- Leptin (116-130) Mouse
- Leptin (93 – 105) Human
- LRRKtide
- MiniAp-4 peptide
- Motilin (1-16)
- Myhc-α 334-352
- Neurokinin A (Substance K)
- Neurokinin B (human, porcine)
- Neuromedin U 25
- Neuropeptide NPSF
- Neuropeptide RFRP-1 (81-92)
- Neuropeptide RFRP-2 (101-112)
- Neuropeptide RFRP-3 (124-131)
- Neuropeptide S human
- Neuropeptide S mouse
- Neuropeptide S rat
- Neuropeptide Y (3-36) Human,Rat
- Neurotensin
- Nociceptin
- NX 210
- Orexin A (monkey)
- Ovalbumin (154-159)
- Ovotransferrin (328-332)
- OXA (17-33)
- Oxidised Alpha-synuclein (1-13)
- Oxytocin (free acid)
- Pep63
- Peptide5
- Polybia-MPII
- SARS-CoV Peptide Antigen negative control
- Spexin
- Substance P
- Thyroglobulin (Tg-FSP)
- Thyroglobulin (Tg-VIF)
- VIP (1-12)
- VIP (6-28)
- [5-FAM]-Galanin (2-30)-[Cys] (Human)
- [Cys]-Galanin (1-30) Human
- [Glp6,Pro9] Substance P (6-11)/Septide
- [Pyr]-Apelin-13
- α-CGRP (mouse, rat)
- Other Categories
- 123B9
- 14-3-3 zeta/delta (28-41)
- 1D,6L-Lanthionine vasopressin
- 3x DYKDDDDK peptide
- a-Gliadin (229-246)
- AD01
- Adrenomedullin (22-52)
- AF10847
- alpha-gliadin (58-73)
- Angiotensin II
- APYTFGQGTK peptide
- Autocamtide-2
- Beclin-1
- Biotin-DAG Peptide
- BMAP-28
- BMF
- BNP-32, porcine
- Bombesin – Potent natural agonist of the mammalian receptor GRPR
- C-telopeptide
- C5aR2 agonist
- cAC 253
- Caloxin 1C2
- Cardiac Targeting Peptide CTP
- CBL (167-180) Light
- CBL (598-612) Light
- CBL-B (22-37) Light
- CBL-B (239-247) Light
- CCK Octapeptide sulfated
- Cecropin-B
- Cellulose synthase 7
- CIGB 300
- Cilengitide
- CNP (1-22), Human, Porcine
- CREB327/active transcription factor CREB-A (113-126) Biotinyl, human
- CREB327/active transcription factor CREB-A (113-126) [5-FAM] amide, Human
- CREB327/active transcription factor CREB-A (113-126), human
- Cysteine Peptide for DPRA tests
- D11-FxxLF Coactivator peptide
- DAG peptide
- Dinitrophenyl ERAP1 peptide
- DYKDDDDK peptide
- Dystrophin (2690-2700)
- Dystrophin (2765-2777)
- Dystrophin (396-405)
- Dystrophin (50-61)
- Dystrophin, DMD
- EHD1
- elf18
- Enfuvirtide (T-20)
- Farnesylated a-factor
- Fas blocking peptide
- FEFEFKFK
- Fibrinogen (377-395) Human
- Flagellin 22 (flg22)
- Formyl-MIFL-acid
- Formyl-Δ-toxin (1-26)
- FSY tripeptide
- Fz7-21
- GALA Peptide
- Ganglioside GM1-binding peptides p3
- Gastrin-1 Rat
- GG-[AMC]
- GIP (1-30) Human amide
- GPRPK pentapeptide
- GQPR tetrapeptide
- GRGDS peptide
- GS dipeptide
- GSS tripeptide
- Heart-homing peptide
- Herceptide
- HiBiT tag
- HIV-1 Rev (34-50)
- HPV16 E6 pep11**m
- HSA (549-558)
- Human Influenza Hemagglutinin (HA) Tag (YPYDVPDYA)
- humanized anti-Tac (HAT) binding peptide
- Hyp3-Bradykinin
- Ig heavy chain V-III region Light
- Ig heavy chain variable region Light
- IGRP Catalytic Subunit-related Protein (206-214)
- Insulin A Chain (A12-21)
- Insulin B (9-23)
- Integral membrane TGN38A (350-361) acetyl, mouse
- Integrin-binding cell adhesive peptide
- Intracellular Sigma Peptide
- JAG-1 (188-204)
- JAG-1, scrambled
- Jelleine 1
- Jelleine 2
- Jelleine 3
- Jelleine 4
- Kallikrein-2 inhibitor
- KLHY-[AMC]
- KR-12-a5 (6-DL)
- KRREILSRRPSYR-acid
- Lasioglossin-III
- LDVP peptide
- Leuprolide Acetate
- LLO (91 – 99)
- Locustatachykinin I
- LP2
- Lys-Glu dipeptide
- Lysine peptide for DPRA tests
- M12 muscle-homing peptide
- M2-Influenza
- MAGEA4 (230-239) Light
- MART-1 (27-35) (human)
- MART-1 Fragment
- Max-1 peptide
- Mouse-ESC-derived cardiomyocyte-targeting peptide
- Mucin 10 (153 – 165), EA2
- MyHC (614-629)
- N-L-Glutamyl-L-Lysine
- Natalizumab (LC46-58)
- Natalizumab LC46-58 KGN deimmunised
- Natalizumab LC46-58 KSN deimmunised
- nef peptide [Human immunodeficiency virus type 1] (73-82) acetyl/amide
- nef protein (75-82) [Human immunodeficiency virus 1]
- nef protein fragment (Acetyl/amide) [Human immunodeficiency virus 1]
- Neuromedin U 8
- Nictide
- Octreotide
- P007 (RXR)4XB
- P12
- P12 amide
- P1A antigen
- Palmitoyl GQPR tetrapeptide
- Palmitoyl KTTKS pentapeptide
- PEN (Mouse)
- PEN, Human
- PEN, Rat
- Pepstatin A
- Pepstatin A Biotin
- Peripheral Myelin Protein P0 (180-199)
- polyalanine peptide (pALA)
- Polybia-MPI
- Polyproline-13
- pp89 phosphoprotein fragment [Mouse cytomegalovirus 1]
- Proinsulin 90-104
- Prolactin-releasing peptide (PrRP20)
- Prostate-specific membrane antigen PSM (35-40), human
- PTD-p65-P1 Peptide
- PUMA BH3
- R5
- Relaxin 3 B1-22R amide
- RGD peptide
- RGD Peptide GRGDSPK
- Rhodopsin Epitope Tag
- S413-PV-[Cys(Npys)]
- SARS-CoV-2 Spike (411-420)
- SARS-CoV-2 Spike (976-984)
- SBCleaner – Peptide decontamination
- Semaglutide Heavy
- SG dipeptide
- SGS tripeptide
- Shepherdin (79 – 87)
- Sialokinins I
- Sifuvirtude
- Skeletal muscle-targeted peptide MSP
- SmBiT
- SMRT peptide
- SOD1 (147-153) human
- Spexin 2 (53-70) Human, Mouse, Rat
- SRC-1 (676-700)
- SSG tripeptide
- Stabilized avi tag peptide
- Steroid Receptor Coactivator-1 (SRC-1) (686-700)
- Suc-LLVY-acid
- Syntide 2
- T-9 peptide
- TAT-GSK’364A
- Teduglutide (GLP2 2G)
- Temporin SHF
- Tet-20
- Tetanus Toxin (1084-1099)
- Tetanus Toxin (1174-1189)
- Tetanus Toxin P30 (947-967)
- Thrombin Receptor Antagonist
- TNF-alpha (1-26), human
- TP10
- TPL-2tide
- Transportan
- TRAP-6 peptide
- Triptorelin acetate
- Truncated flagellin 22 (flg22)
- Tumstatin (69-88)
- Ub4ix
- UBA3 (59-72) peptide
- Urumin
- V5 epitope tag
- Vasculotide
- VGB4
- VIP (guinea pig)
- Visperas1pY
- Visperas2pY
- Xenin
- YSA amide
- [5-FAM]-DAG peptide
- [Azhx]-ANP (Human)
- [Cys] citrullinated alpha enolase
- [Cys]-Exendin 4
- [Nle12] a-factor
- [Tyr]-CNP22, Human
- εV1-2
- Protein-Protein Interactions
- Stable Isotope Labeled (SIL) peptides catalog
- ADALQAGASQFETSAAK* – SIL Infliximab signature peptide
- APYTFGQGTK* – SIL Adalimumab internal standard
- ASGYTFTSYNMHWVK* – SIL Rituximab signature peptide quantifier
- ASQSIGTNIHWYQQR* – SIL Cetuximab signature peptide qualifier
- DYAMTWVR* – SIL Dupilumab signature peptide qualifier
- GLEWIGAIYPGNGDTSYNQK* – SIL Rituximab signature peptide qualifier
- Heavy Angiotensin II
- Heavy calcitonin peptide
- LEWIGEIDPSESNTNYNQK* – SIL Vedolozumab signature peptide
- LSITIRPR* – SIL Dupilumab signature peptide quantifier
- NYLAWYQQKPGK* – SIL Adalimumab signature peptide quantifier
- QAPGQGLEWMGDINTR* – SIL Emicizumab signature peptide qualifier
- SGGSIYNEEFQDR* – SIL Emicizumab signature peptide quantifier
- SINSATHYAESVK* – SIL Infliximab signature peptide quantifier
- SLEWIGAIDPYYGGTSYNQK* – SIL Dinutuximab signature peptide qualifier
- SSSTAYMHLK* – SIL Dinutuximab signature peptide quantifier
- YASESISGIPSR* – SIL Cetuximab signature peptide quantifier
- YASESMSGIPSR* – SIL Infliximab signature peptide qualifier
- TAT Conjugated Peptides
- Cys-TAT(48-60)
- dfTAT
- fTAT
- gp91 ds-TAT
- SMAC/DIABLO -TAT (48-60)-[Lys]
- TAT (48-57)
- TAT (48-59) amide
- TAT (48-60) amide
- TAT – GluR23Y
- TAT 2-4
- TAT protein (28-35) [Simian immunodeficiency virus]
- TAT-AKAP79 (326-336) amide
- TAT-AKAP79 (326-336) scrambled
- TAT-AKAP79 (326-336) scrambled amide
- TAT-Beclin 1
- TAT-Beclin Scrambled
- TAT-CHN9 (C-ter)
- TAT-CN21
- TAT-NR2B (C-ter)
- TAT-Pro ADAM10 (709-729)
- TAT-TRPV1 (736-745)
