ALPHA FACTOR SIGNALING PEPTIDE
Yeast mating signaling pathway
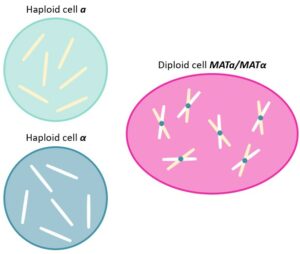
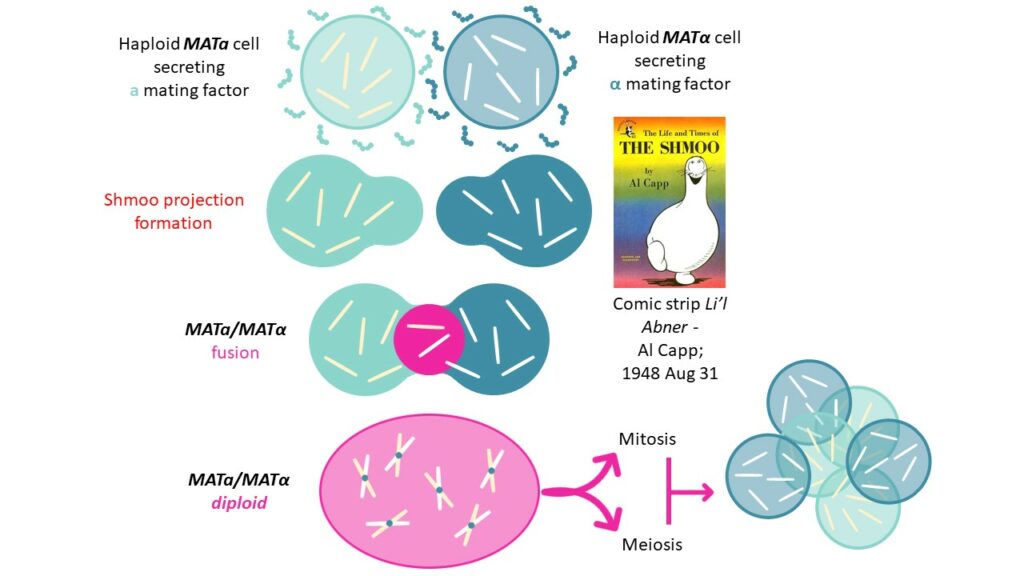
The life cycle of Saccharomyces cerevisiae,also known as the brewer’s and the baker’s yeast,is characterized by both haploid and diploid phases. Indeed,yeasts are organized in two haploid types,known as a and α cells (MATa and MATα),that can either proliferate individually,using mitosis,or fuse together to form MATa/MATα diploids.
For two haploid cells to mate,mating factors are needed. These pheromones induce G-Protein Coupled Receptor‘s (GPCR) conformational changes,which allow G-protein migration in the plasma membrane. GPCRs are present on both a and α yeast cells and turn into Guanine nucleotide Exchange Factors (GEF) upon G-protein release. The receptor’s « new activity » leads to guanosine diphosphate (GDP) exchange by guanosine triphosphate (GTP) on the G-protein,which will recruit STE 5 to allow Fus3 protein phosphorylation. Phosphate addition on Fus3 enables its fusion to the cell’s plasma membrane in order to activate formin proteins by phosphorylation.
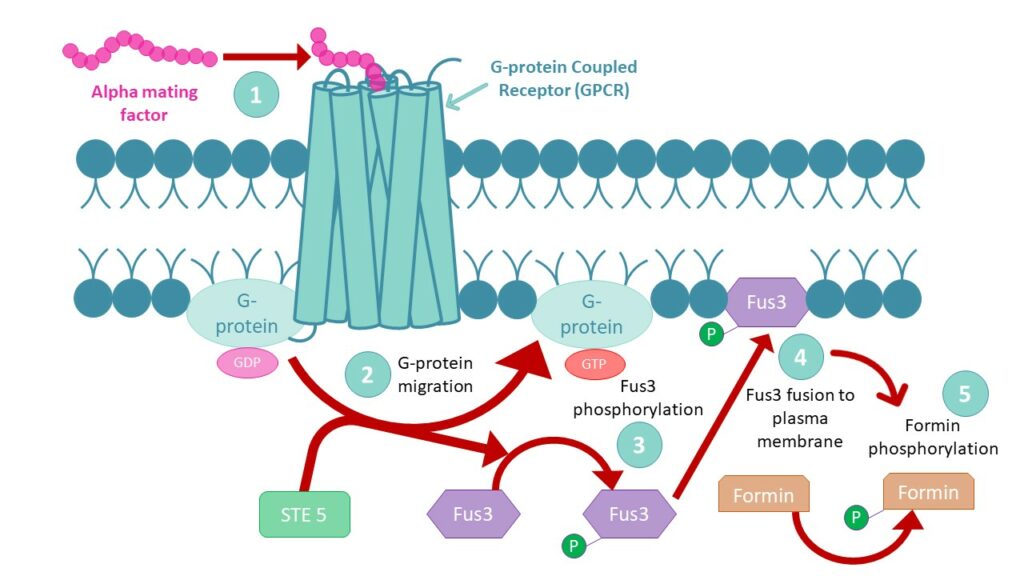
Formin proteins being involved in the polymerization of microfilaments,their consequent growth within the cell will form shmoo projections to enable complete mating of MATa and MATα cells,towards MATa/MATα diploid cell that can undergo mitosis and meiosis to create new haploid spores.
[CAS: 59401-28-4] α-factor pheromone – WHWLQLKPGQPMY
To induce a yeast cells’ shmoo projection,WHWLQLKPGQPMY pheromone (or alpha mating factor) is produced by α yeast cells. This signaling peptide is part of the MFAL1_YEAST protein (UniProt: P01149) that excretes its active factor into the culture medium to act on the opposite mating type).
α mating factor is a 13 residues peptide inducing the expression of necessary mating genes ( 200 genes – 3% of the yeast’s genome) to arrest yeast cell cycle in the G1 phase,while altering cell surface and nuclear determinants,towards MATa and MATα fusion.
WHWLQLKPGQPMY interacts with its complementary pheromone,a-factor,which is a 12 residues peptide (YIIKGVFWDPAC) that is covalently attached to farnesyl to generate the appropriate response upon α pheromone activation.
Alpha factor applications
SB-PEPTIDE offers α mating factor WHWLQLKPGQPMY to study:
- Cell cycle
- Cellular morphology
- Transcriptional induction
- Signal transduction pathways
Technical specification
 |
Sequence : WHWLQLKPGQPMY |
 |
MW : 1683 ,97 g/mol (C82H114N20O17S) |
 |
Purity : > 95% |
 |
Counter-Ion : TFA Salts (see option TFA removal) |
 |
Delivery format : Freeze dried in propylene 2mL microtubes |
 |
Other names : Alpha factor,Alpha mating pheromone,α-factor,α pheromone |
 |
Peptide Solubility Guideline |
 |
Bulk peptide quantities available |
Price
| Product catalog | Size | Price € HT | Price $ USD |
| SB282-5MG | 5 mg | 220 | 275 |
| SB282-50MG | 50 mg | 605 | 756 |
| SB282-100MG | 100 mg | 1210 | 1513 |
References
Genetics. 2014 May;197(1):33-48. doi: https://doi.org/10.1534%2Fgenetics.114.163188
Budding Yeast for Budding Geneticists: A Primer on the Saccharomyces cerevisiae Model System
The budding yeast Saccharomyces cerevisiae is a powerful model organism for studying fundamental aspects of eukaryotic cell biology. This Primer article presents a brief historical perspective on the emergence of this organism as a premier experimental system over the course of the past century. An overview of the central features of the S. cerevisiae genome,including the nature of its genetic elements and general organization,is also provided. Some of the most common experimental tools and resources available to yeast geneticists are presented in a way designed to engage and challenge undergraduate and graduate students eager to learn more about the experimental amenability of budding yeast. Finally,a discussion of several major discoveries derived from yeast studies highlights the far-reaching impact that the yeast system has had and will continue to have on our understanding of a variety of cellular processes relevant to all eukaryotes,including humans.
Genes Dev. 2002 Jul 1;16(13):1587-609. doi: https://doi.org/10.1101/gad.1003302
Guanine nucleotide exchange factors for Rho GTPases: turning on the switch
Rho GTPases control many aspects of cell behavior through the regulation of multiple signal transduction pathways (Van Aelst and D’Souza-Schorey 1997; Hall 1998). Rho,Rac,and Cdc42 were first recognized in the early 1990s for their unique ability to induce specific filamentous actin structures in fibroblasts; stress fibers,lamellipodia/membrane ruffles,and filopodia,respectively (Hall 1998). Over the intervening years,evidence has accumulated to show that in all eukaryotic cells,Rho GTPases are involved in most,if not all,actin-dependent processes such as those involved in migration,adhesion,morphogenesis,axon guidance,and phagocytosis (Kaibuchi et al. 1999; Chimini and Chavrier 2000; Luo 2000). In addition to their well-established roles in controlling the actin cytoskeleton,Rho GTPases regulate the microtubule cytoskeleton,cell polarity,gene expression,cell cycle progression,and membrane transport pathways (Van Aelst and D’Souza-Schorey 1997; Daub et al. 2001;Etienne-Manneville and Hall 2001). With such a prominent role in so many aspects of cell biology,it is not surprising that they are themselves highly regulated…
Mol Cell Biol. 1996 Sep;16(9):4700-9. doi: https://doi.org/10.1128%2Fmcb.16.9.4700
Yeast α Mating Factor Structure-Activity Relationship Derived from Genetically Selected Peptide Agonists and Antagonists of Ste2p
α-Factor,a 13-amino-acid pheromone secreted by haploid α cells of Saccharomyces cerevisiae,binds to Ste2p,a seven-transmembrane,G-protein-coupled receptor present on haploid a cells,to activate a signal transduction pathway required for conjugation and mating. To determine the structural requirements for α-factor activity,we developed a genetic screen to identify from random and semirandom libraries novel peptides that function as agonists or antagonists of Ste2p. The selection scheme was based on autocrine strains constructed to secrete random peptides and respond by growth to those that were either agonists or antagonists of Ste2p. Analysis of a number of peptides obtained by this selection procedure indicates that Trp1,Trp3,Pro8,and Gly9 are important for agonist activity specifically. His2,Leu4,Leu6,Pro10,a hydrophobic residue 12,and an aromatic residue 13 are important for both agonist and antagonist activity. Our results also show that activation of Ste2p can be achieved with novel,unanticipated combinations of amino acids. Finally,the results suggest the utility of this selection scheme for identifying novel ligands for mammalian G-protein-coupled receptors heterologously expressed in S. cerevisiae.
Cell. 1983 Mar; Vol 32(3):P839-852. doi: https://doi.org/10.1016/0092-8674(83)90070-3
Yeast α factor is processed from a larger precursor polypeptide: The essential role of a membrane-bound dipeptidyl aminopeptidase
Alpha factor mating pheromone is a peptide of 13 amino acids secreted by Saccharomyces cerevisiae α cells. Nonmating (“sterile ,” or ste) α-cell mutants bearing defects in the STE13 gene do not produce normal α factor,but release a collection of incompletely processed forms (α factor∗) that have a markedly reduced specific biological activity. The major α-factor∗ peptides have the structures H2N-GluAlaGluAla-α factor and H2N-AspAlaGluAla-α factor. The ste13 mutants lack a membrane-bound heat-stable dipeptidyl aminopeptidase (DPAPase A) that specifically cleaves on the carboxyl side of repeating -X-Ala- sequences. Absence of DPA-Pase A and the other phenotypes of a ste13 lesion cosegregate in genetic crosses. The cloned STE13 gene on a plasmid causes yeast cells to overproduce DPAPase A severalfold. A different cloned DNA segment,which weakly suppresses the ste13 defects,causes overproduction of a heat-labile activity (DPAPase B) by about tenfold. Other experiments indicate that DPAPase A action may be ratelimiting for α-factor maturation in normal α cells.
